How SciDAP works
SciDAP is a no-code bioinformatics platform
that enables scientists to analyze NGS-based data
without a bioinformatician or any knowledge of coding.
Overview
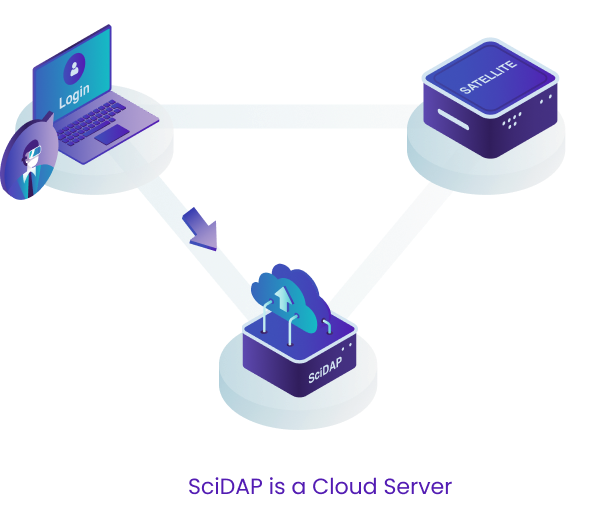
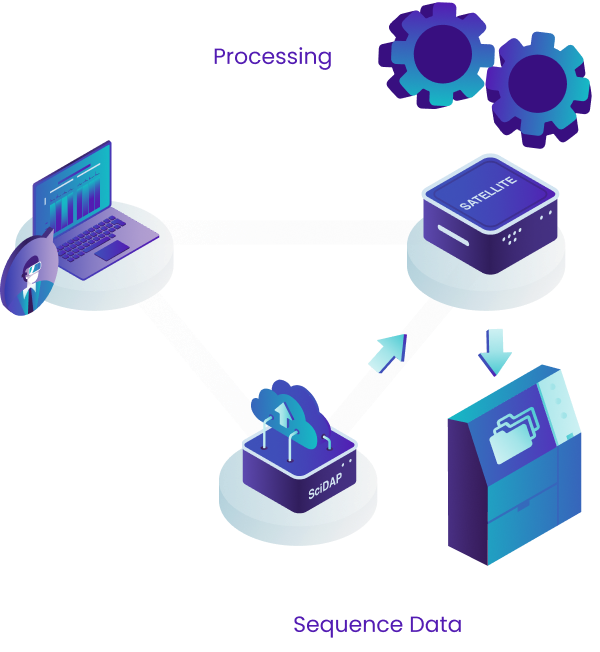
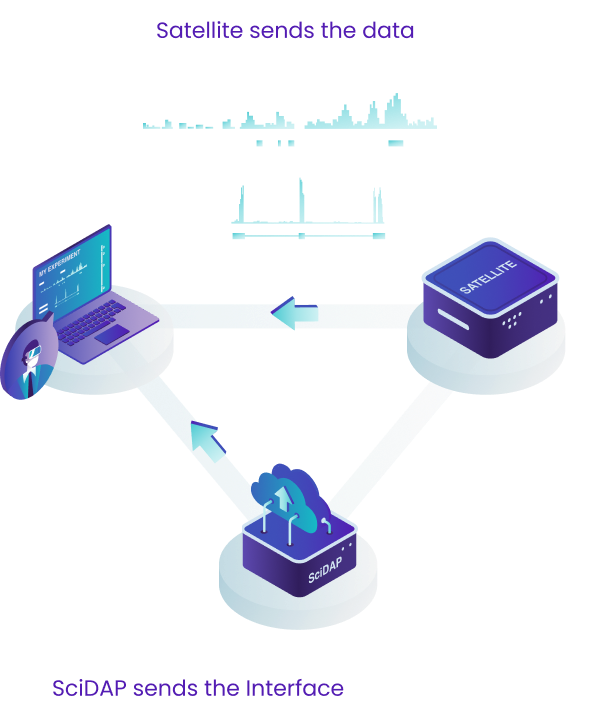
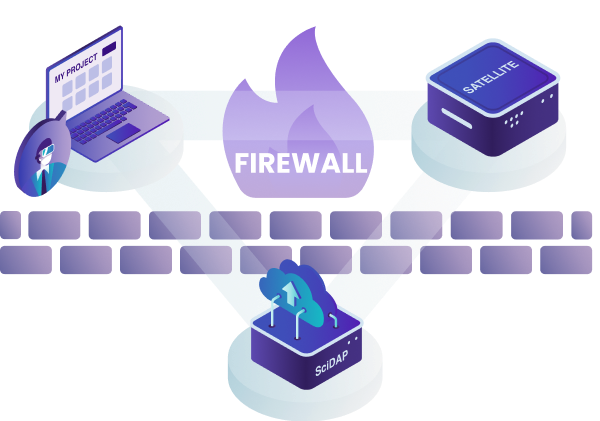
SciDAP is a cloud enabled bioinformatics analysis platform that provides an intuitive user interface and seamless data processing and storage.
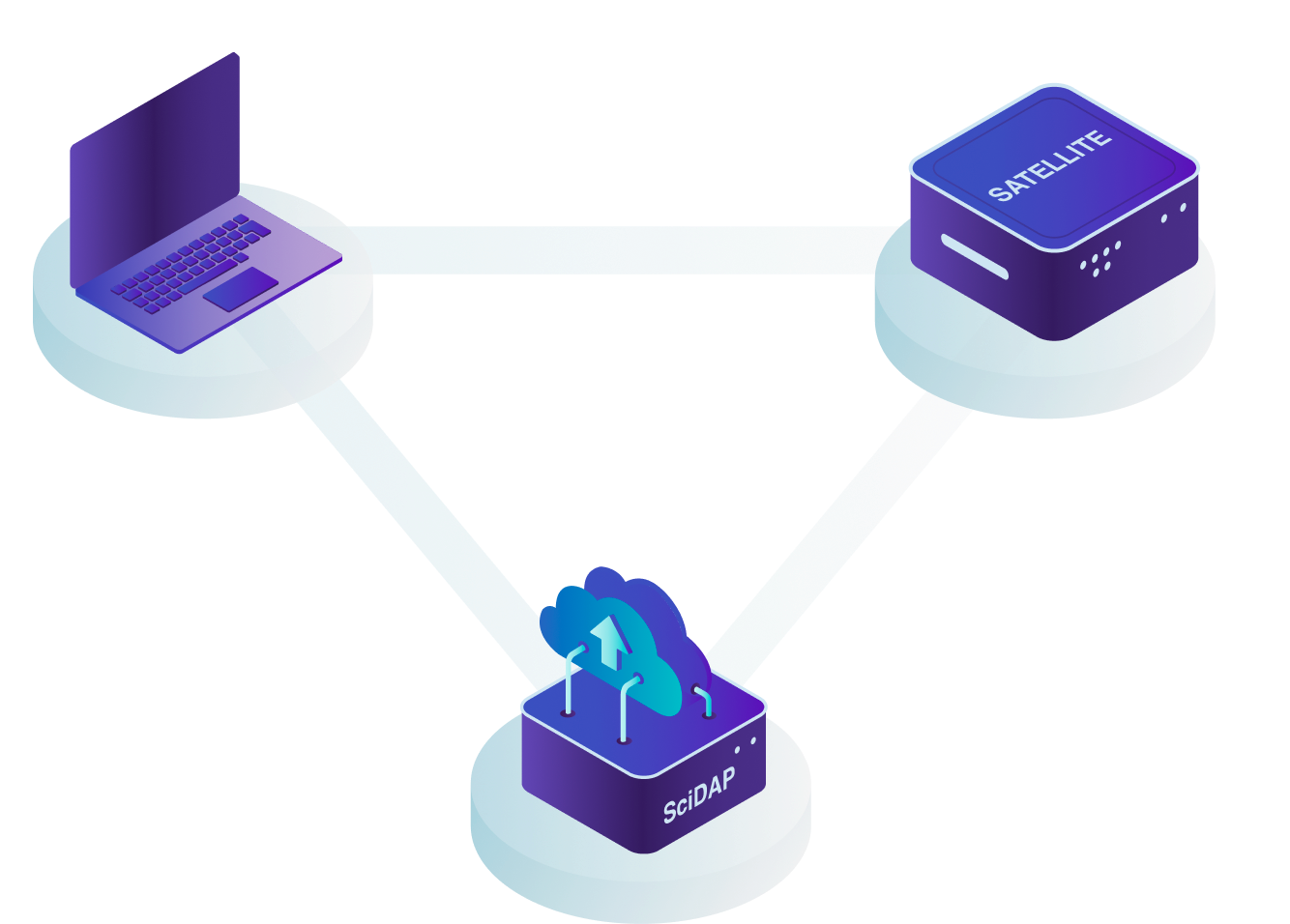
This design allows us to provide users with the most up-to-date interface and to quickly deploy new pipelines through the central cloud server. If, however, there are HIPAA or data privacy needs, we also provide a "satellite" solution where the data never has to leave your network. This is described in more detail below, just keep scrolling!
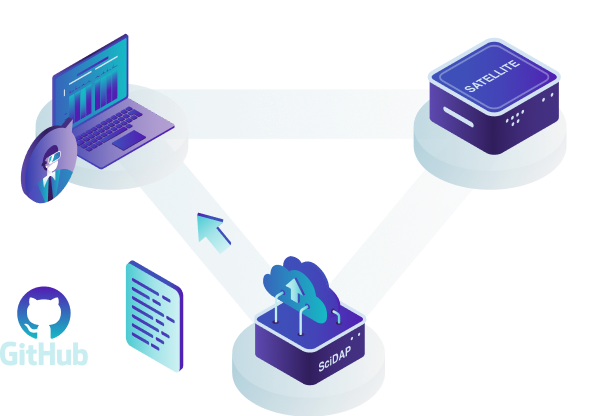
For either approach, users can access SciDAP from any computer by logging into scidap.dev. The SciDAP platform is designed to scale to your needs, and can serve as a data processing and exchange center for your whole laboratory or institution. All authorised laboratory members can add data, execute analyses, and view and share results.
 scidap
scidap