CUT&RUN and CUT&TAG
Overview
CUT&RUN and CUT&Tag are next-generation sequencing (NGS) applications that identify the DNA sequence where DNA binding proteins bind DNA in chromatin. Developed in the Henikoff Laboratory at Fred Hutchinson Cancer Center, they provide an alternative to ChIP-Seq. CUT&RUN and CUT&Tag offer advantages over ChiP-Seq because they work with fewer cells and eliminate the crosslinking that occurs outside of the cell, helping to retain cellular context.
General Workflow
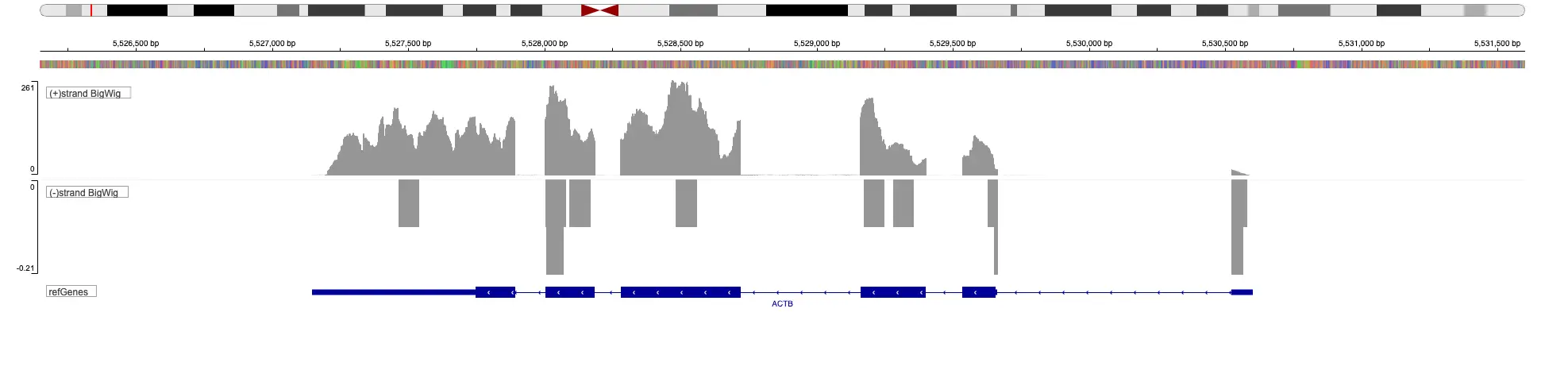
CUT&RUN and CUT&Tag begin by permeabilizing the cells and the addition of an antibody specific for your protein of interest. After the binding and wash steps, a fusion protein composed of protein A/G with either micrococcal nuclease or Tn5 transposase is added. This fusion protein binds to the antibody via the protein A/G domain. After a wash step, calcium or magnesium is added to activate enzymatic activity. For CUT&RUN, micrococcal nuclease will cut DNA near the binding site. For CUT&Tag, the transposase will insert specific oligos next to the sites. The DNA is subsequently amplified for CUT&Tag or after the addition of adapters for CUT&RUN. The released DNA is sequenced to identify the protein binding sites.
Data Analysis
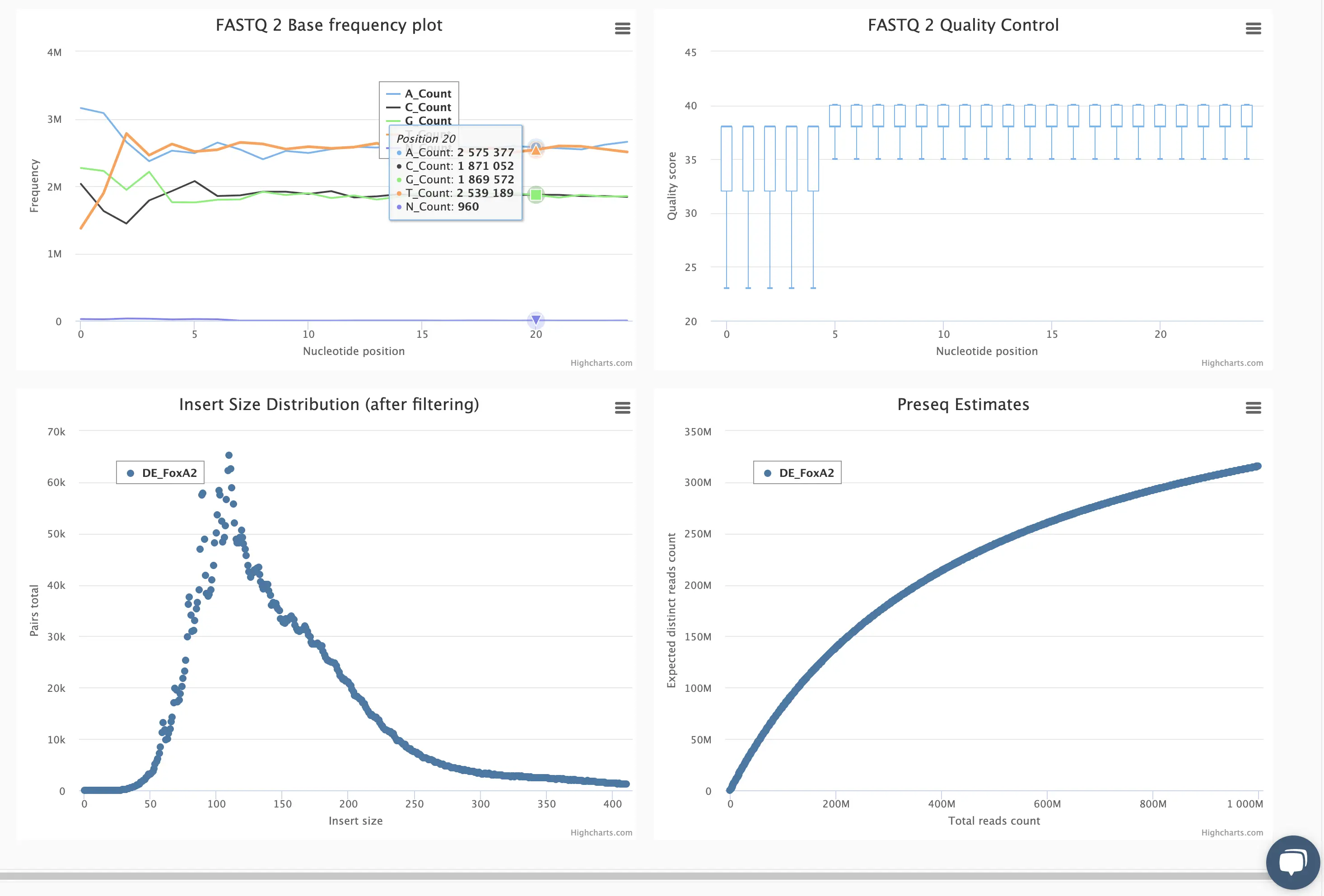
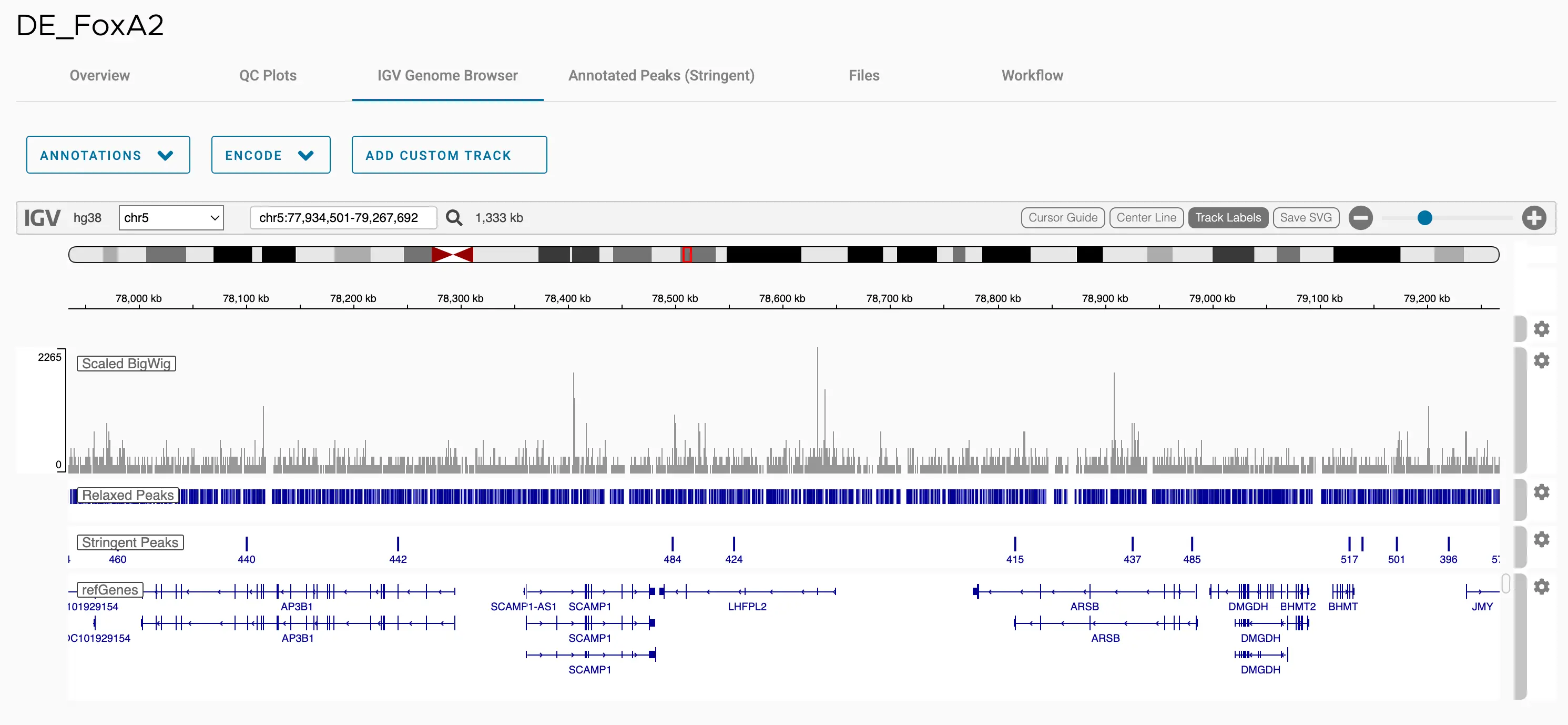
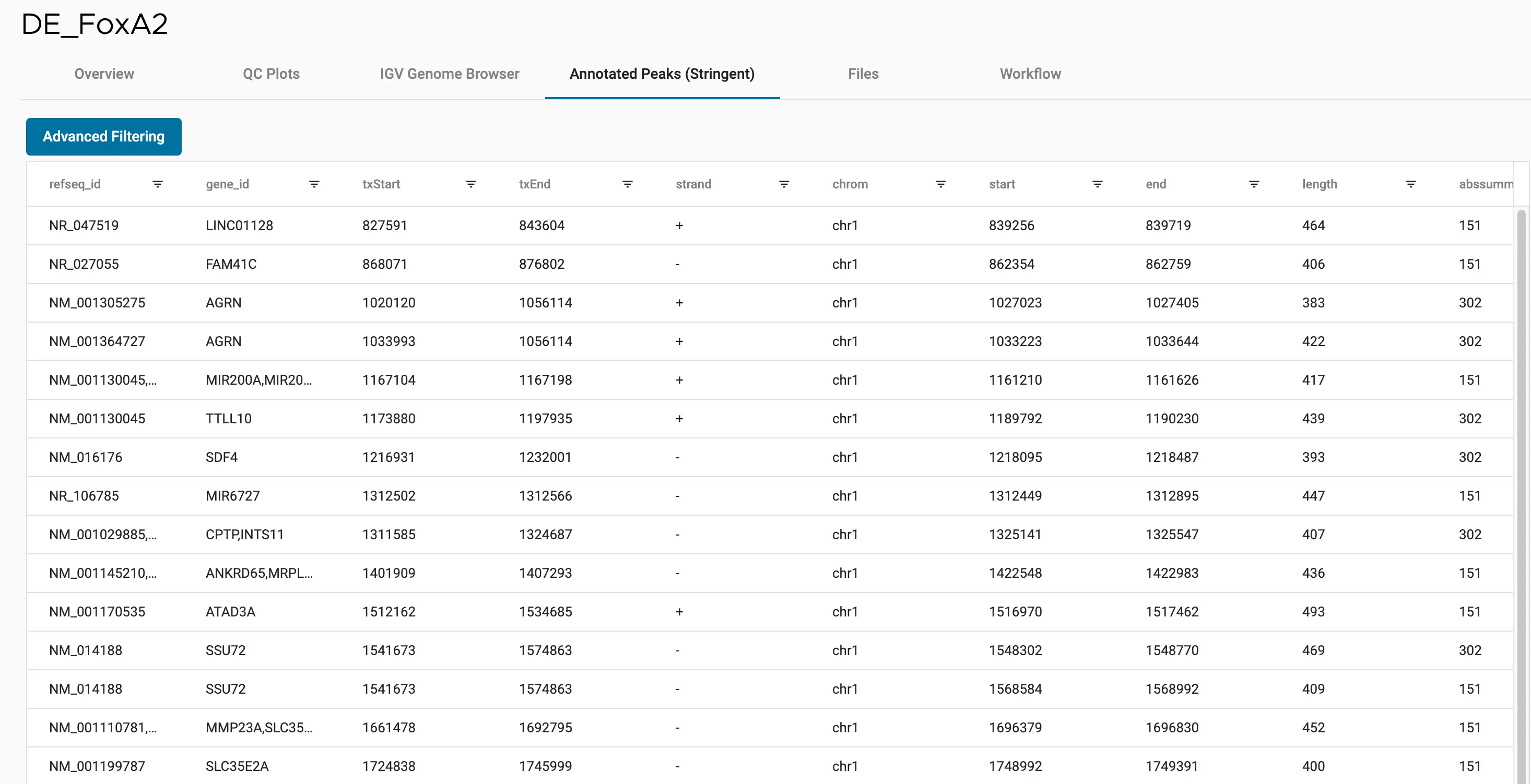
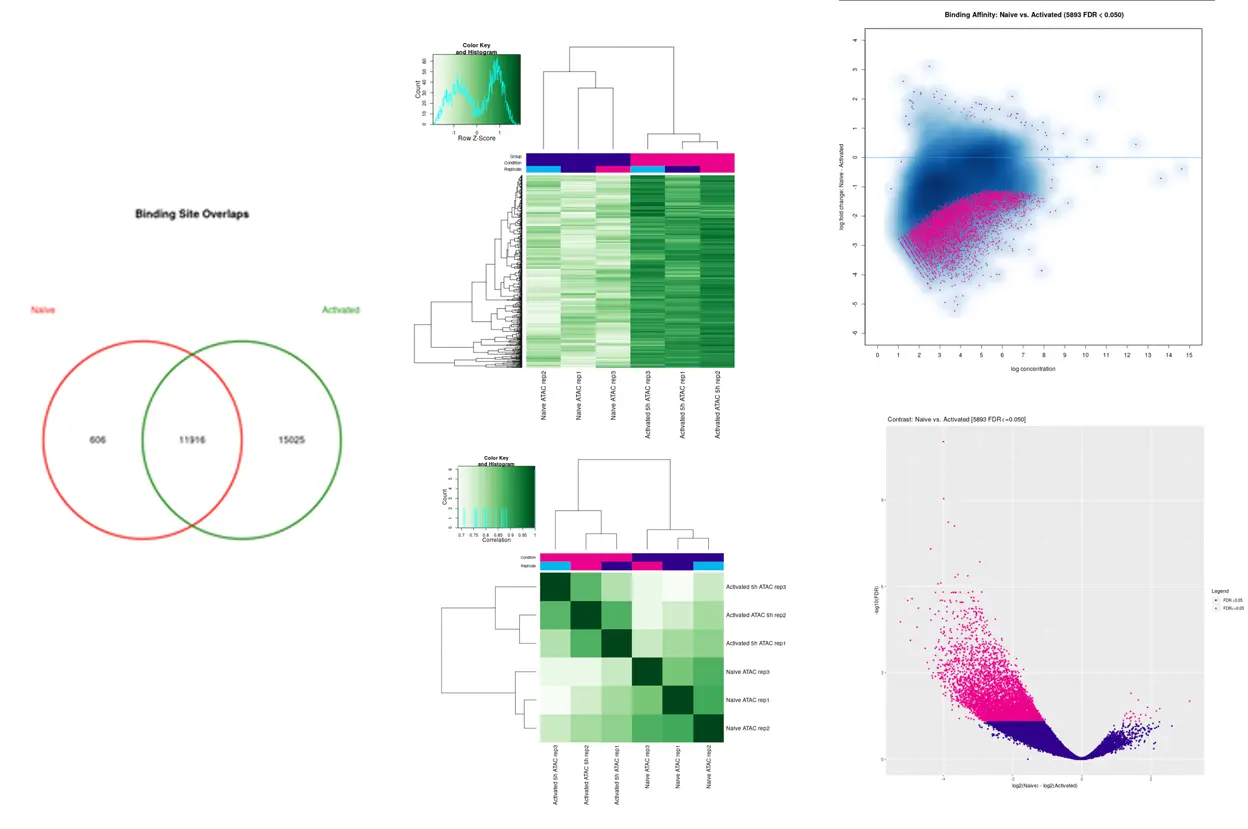
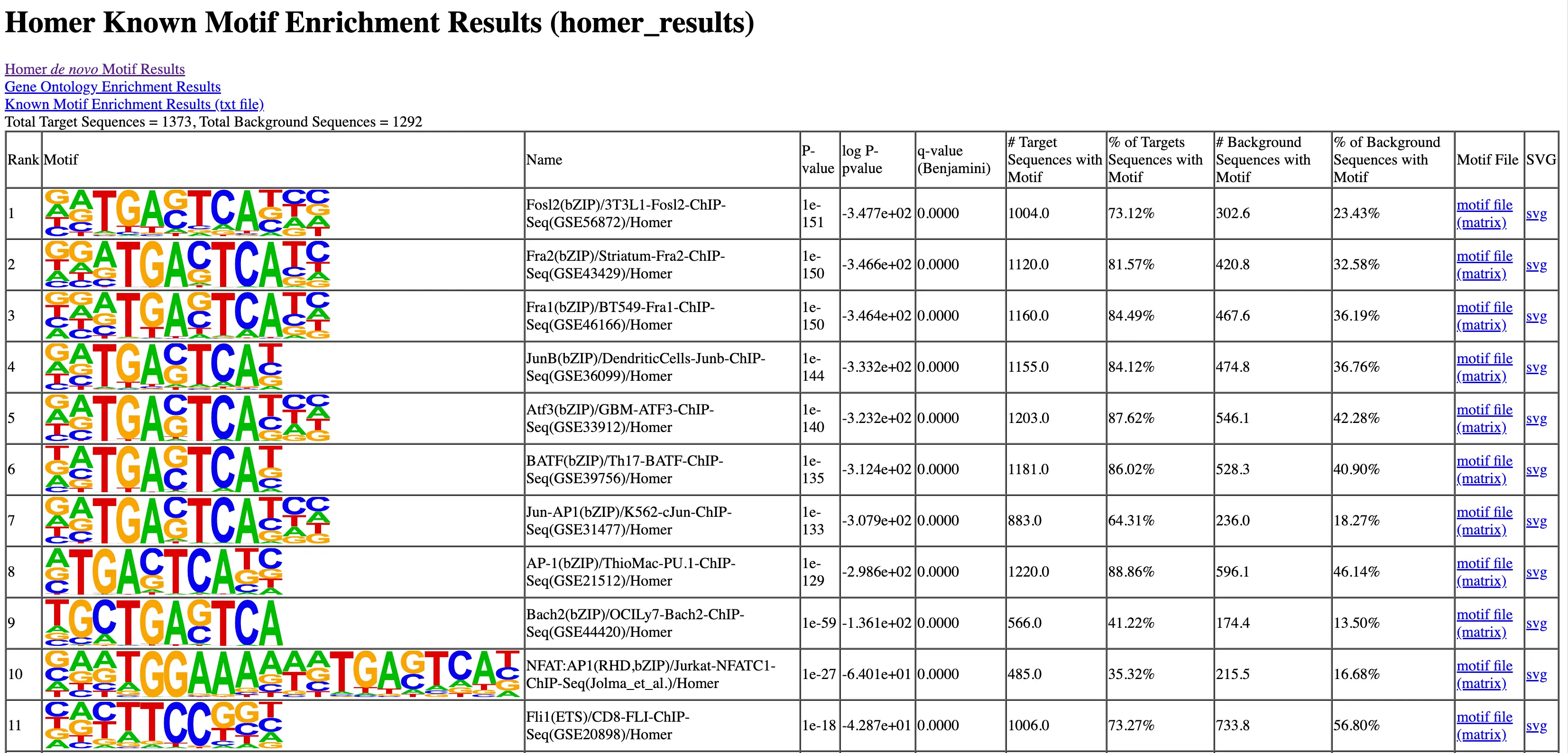
SciDAP is a no-code bioinformatics platform that enables scientists to analyze NGS-based data without a bioinformatician. It has built-in pipelines based on open-source workflows to analyze bulk data from CUT&RUN and CUT&TAG libraries. SciDAP starts from FASTQ data files from a paired-end (PE) CUT&RUN or CUT&Tag sequencing library provided by most DNA core facilities and commercial service providers.
 scidap
scidap